Objectives:
Mutations in the NPM1 gene are found in more than 30% of acute myeloid leukaemia (AML) cases. The mutations disrupt a nucleolar localization signal (NoLS) and create a novel nuclear export signal (NES), leading to cytoplasmic displacement of the protein (NPM1c). NPM1cmutations prime haematopoietic progenitors to leukaemic transformation, but their precise molecular consequences remain elusive. Here, we first examine the effects of isolated NPM1c mutations on mouse haematopoietic cells and explore their impact on their transcriptome, including at single-cell resolution, and their proteome. Then, using several murine and human AML models and datasets we investigate the role of such NPM1c-associated changes in fully transformed AML cells and explore their potential as therapeutic vulnerabilities.
Methods:
We performed RNA-seq, CITE-seq and TMT global quantitative proteomics using lineage-depleted bone marrow progenitor cells from NPM1 flox-cA/+;Mx1-cre mice ( Vassiliou et al, Nat Genet, 2011). Proteomic changes were contrasted to those identified in a recently published dataset of 44 human AMLs, including 8 NPM1-mutant AMLs. Candidate vulnerabilities were identified using datasets from CRISPR-Cas9 drop-out screens and were validated in NPM1 cA/+-harbouring cell lines and primary mouse NPM1 cA/+;FLT3 ITD/+ AMLs.
Results:
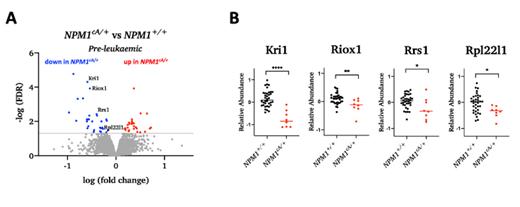
We found a global increase in the protein abundance of several members of the KPN family in pre-leukaemic Npm1 cA/+ cells, an effect also observed in human NPM1c+ AMLs. Moreover, we observed a decrease in the relative abundance of several ribosome biogenesis-related proteins in pre-leukaemic Npm1 cA/+ cells and confirmed that this effect persists in human NPM1c+ AML patient samples. We go on to show that, when compared to NPM1 +/+ cells, pre-leukaemic NPM1 cA/+cells show high sensitivity to Actinomycin D and other RNA pol I inhibitors, but not to the translation inhibitor cycloheximide. Combination treatment with Actinomycin D and Venetoclax inhibited the growth and colony forming ability of both pre-leukaemic NPM1 cA/+cells and primary mouse Npm1 cA/+;Flt3 ITD/+AMLs. By contrasting our findings with data from CRISPR drop-out screens, including a genome-wide screen in primary mouse Npm1 cA/+;Flt3 ITD/+/Cas9AMLcells, we identified and validated ribosome biogenesis factors, such as Tsr3, whose knock-out selectively affected cell growth, survival and clonogenicity of NPM1 +/+AML cells, therefore representing novel potential therapeutic targets that could be exploited in the clinic.
Conclusions:
We show that NPM1c mutations are associated with reduced levels in several ribosome biogenesis factors resulting in therapeutic vulnerabilities of NPM1-mutant pre-leukaemic and leukaemic (AML) cells. The development of novel therapeutics to target these factors may help improve the survival of patients with NPM1-mutant AML.
Figure Legend: (A) Volcano plot showing protein abundance in mouse NPM1 cA/+vs NPM1 +/+ pre-leukaemic haematopoetic progenitors. (B) Relative abundance of ribosome-related proteins in human NPM1 cA/+vs NPM1 +/+AML samples (dataset from Kramer et al., 2022).
Disclosures
Vassiliou:AstraZeneca: Other: Educational Grant; STRM.BIO: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal